Exploring the PCR Assay for Detecting Tropomyosin: Major Allergen in Shrimp-Derived Ingredient in Food
Nam Hoang Dang Phan, Tung Thanh Nguyen, Tran Bao Hoang Tran, Nhan Thanh Vo, Trinh Tuyet Thi Le, Minh Trong Quang, Thuy Ai Huyen Le, Thuan Duc Lao*
|
|
Faculty of Biotechnology, Ho Chi Minh City Open University, Ho Chi Minh City, Vietnam. |
ABSTRACT
Shrimp is the popular food within high nutritional values. Specially, the shrimp is consumed as a raw material in various processed foods worldwide. However, shrimp can commonly trigger the seafood allergic reactions that threaten the lives of people. In this study, the PCR assay was developed to detect the presence of tropomyosin - a shrimp-derived major allergen. For allergen detection, a specific primer pair, Tropo-F and Tropo-R, was designed based on the shrimp tropomyosin gene. The specificity of primer was accessed by in silico study and initially applied in a sample of Litopenaeus vannamei and commercial shrimp-derived processed food. As the results, an 81-bp band was observed in the sample of Litopenaeus vannamei and other commercial shrimp-derived component foods, and no band was observed in non-shrimp samples. The specificity was confirmed by sequencing, which indicated that the PCR products were homologous to Litopenaeus vannamei and Penaeus monodon. In summary, these results indicated that PCR assay with Tropo-F and Tropo-R primers could successfully identify the tropomyosin in processed food, and could be applied to a large number of samples in the reality. To increase the value of this PCR assay, the sensitivity of this assay is necessary to be evaluated in our near studies.
Keywords: tropomyosin, food allergy, shrimp-derived processed food
Introduction
Shrimp belongs to the group of crustaceans (Anthropods), is the popular food that is rich in nutritional value within Vitamins, Minerals as well as proteins [1-4]. Additionally, shrimp is consumed as a raw material in various processed foods such as shrimp seasoning, fried snack foods, etc [5]. In reality, shrimp and shrimp-derived ingredient product are the common and commercial foods worldwide. However, shrimp has been considered as one of the major causes of seafood allergies [1, 6]. The prevalence of shrimp allergies is not reported in every country. However, the clinical reports of shrimp allergies are frequently reported [7]. The symptoms of crustaceans include shrimp allergy, commonly present with urticarial, flushing, itching, as well as respiratory system problems such as dyspnea, wheezing, etc. that seriously affect the health of consumers. Seriously, allergic reactions could lead to anaphylactic shock that threatens the lives of people [2, 8]. Shanti et al. (1993) identified the major heat-stable allergen of shrimp, tropomyosin, which is capable of provoking the hypersensitivity reactions due to the intake of shrimp [9]. Later, tropomyosin was also identified as the allergen in shellfish, comprised of the group of crustaceans, including crabs, prawns, shrimps, and a group of mollusks, including bivalves, gastropods, and cephalopods [10, 11]. Therefore, tropomyosin is identified as the molecular target for the identification of allergen in shrimps, shrimp-derived ingredient foods and other shellfish foods. Although the processed foods are undertaken by the heat or pressure, DNA has been identified as the stable molecule in processed conditions [12], therefore, compared to the protein-based methods, DNA-based method is more suitable in detecting food allergens [11, 13]. Therefore, it is important to establish a PCR-method to detect the allergen in shrimp and shrimp-derived ingredients in foods. In the current study, the designed primers and PCR assay were established based on their tropomyosin gene to identify the allergen in shrimp-derived components in food.
Materials and Methods
Designed primers
The tropomyosin gene of various kinds of shrimp was collected from Genbank (NCBI) with different accession numbers, including MH999806 (Penaeus setiferus), AB270629 (Penaeus monodon), EU410072 (Litopenaeus vannamei), XM_027373346 (Penaeus vannamei), GU233303 (Fenneropenaeus chinensis), etc. The alignment of various tropomyosin genes was performed by BioEdit Sequence Alignment Editor Software (https://bioedit.software.informer.com/). The primers were artificially designed based on the consensus of tropomyosin sequences. Then, the characteristic and specificity of primers were evaluated by using IDT oligo analyze as well as primer-BLAST.
Sample collection, DNA extraction and PCR assay
The sample of Penaeus vannamei as the positive control, as well as shrimp-derived ingredient foods and raw meat (Pork) used as controls, were collected and enrolled in the current study. The total of DNA was extracted from 1g of samples using Phenol (pH = 8)/Chloroform method. The DNA quantity was checked by evaluating the absorbance at OD260 and OD280. The pure DNA with OD260/OD280 ratio of 1.8–2.0 was used for further PCR assay.
The PCR was carried out for the samples with a total volume of 15 µL containing 250 ng DNA template, and PCR Master mix 2X (Thermo Fisher Sciencetific). PCR reaction was subjected to initial incubation at 95oC for 5 min, followed by 40 cycles at 95oC for 30 s, 53oC for 30 s, 72oC for 30 s, and 72oC for 5 min for the final incubation. The PCR products were directly loaded onto a 2.0% agarose gel, stained with ethidium bromide, and directly visualized under ultraviolet illumination. Then, the PCR products were sequenced to confirm the specificity of primers.
Results
The designed primers: Tropo-F and Tropo-R
The Trop-F (Forward primer) and Tropo-R (Reversed primer) sequences were used to amplify the tropomyosin gene region based on the tropomyosin gene collected from various kinds of shrimp in Genbank. The sequences and characteristic of Tropo-F and Tropo-R are shown in Table 1. As shown in Table 1, the characteristics of Tropo-F and Tropo-R including length, melting temperature, %GC content, avoiding the secondary structures, etc. were proper and suitable for the successful PCR reaction. The result of the alignment of the primers and various kinds of shrimp tropomyosin gene showed that the 81-bp PCR product was amplified by using the pair of Tropo-F and Tropo-R (Fig. 1). Based on the BLAST results, only the trypomyosin gene was amplified, which indicated specificity. Therefore, we concluded that the trypomyosin-amplified primers were successfully designed.
Table 1. The sequences and characteristic of Tropo-F and Tropo-R used in amplifying the gene of tropomyosin.
|
Primer |
Sequence 5’ – 3’ |
L (bp) |
Tm (oC) |
%GC |
(1) |
(2) |
(3) |
|
Tropo-F |
GTTGGTTGAGCACCTCCT |
18 |
54.9 |
55.6 |
-1.45 |
-4.41 |
-5.02 |
|
Tropo-R |
ATCCTTCTCCAGCTTCATCGC |
21 |
57.1 |
52.4 |
0.41 |
-6.43 |
|
Note: L: Length; Tm: Melting temperature; Gibbs free energy (kcal/mole) for (1) hairpin loop; (2) selfdimer; and (3) heterodimer.

Figure 1. The primers Tropo-F and Tropo-R matched on the alignment of various kinds of shrimp tropomyosin gene, which yielded the PCR product with the length of 81 bps.
PCR assay with Tropo-F and Tropo-R applied in the detection of tropomyosin in commercial food
The samples of Penaeus vannamei, shrimp-derived ingredient foods, and non-shrimp samples (Pork) were collected from the local supermarket and used in the current study. The presence of tropomyosin was analyzed by PCR assay with the primer set of Tropo-F and Tropo-R, as shown in Fig. 2. As a result, only one 81-bp band was observed in the sample of Penaeus vannamei and shrimp-derived ingredient foods. In the case of the control sample and negative control, no amplifier result was observed. Sequencing was done to confirm the primers’ specificity and the 81-bp-length products (yielded by Tropo-F and Tropo-R primer) were observed as shown in Fig. 3. According to results, the signals of peaks in PCR products sequencing were good for nucleotide reading. The results of the BLAST of both sequencings were summarized in Table 2. The amplified sequences, yielded by Tropo-F and Tropo-R primer, generated in our study revealed in Penaeus vannamei and Penaeus monodon samples, respectively. Therefore, the results that confirmed PCR assay with Tropo-F and Tropo-R primers' amplification in this study could be applied in detection of tropomyosin gene in foods.
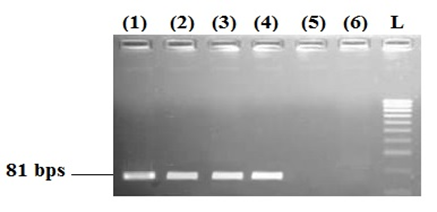
Figure 2. The electrophoresis of the samples' PCR products. (1) Penaeus vannamei; (2)-(4); shrimp-derived ingredient foods; (5) pork – control sample; (6) negative control.
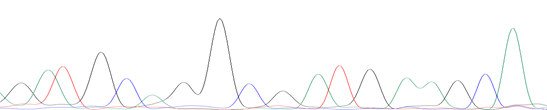
(A)
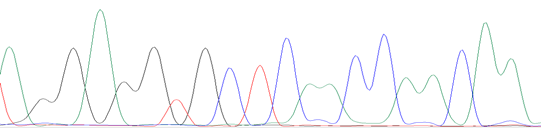
(B)
Figure 3. The represented sequence identified tropomyosin in (A) Penaeus vannamei sample; (B) shrimp-derived ingredient food (No. well 2 product)
Table 2. BLAST results of the sequencing of represented amplifiers
|
Sequencing samples |
BLASTn (Genbank) |
|||
|
Homology |
Accession number |
Homology rate |
E-value |
|
|
(1) |
Penaeus vannamei |
XM_027373346 |
100.00% |
0.1 |
|
(2) |
Penaeus monodon |
AY827100 |
86.11% |
9e-05 |
|
(3) |
Penaeus vannamei |
XM_027370375 |
34.60% |
0.1 |
|
(4) |
Penaeus vannamei |
XM_027373345 |
100.00% |
0.1 |
Discussion
Up to date, a variety of methods have been used to detect food allergens worldwide. The main commercial assay for detecting food allergens is protein-based assays, such as enzyme-linked immunosorbent assays (ELISAs). The protein-based assays have advantages; for example they are easy to use, detectable in a short period of time, as well as directly react with allergen-inducing antigens [14]. However, the limitation of the protein-based assay is the low sensitivity of the high-temperature processed foods, resulting in allergic proteins to be denatured. Therefore, DNA-based assays have been developed so far. In the case of DNA-based assays, the DNA molecule was identified as the thermal-stable and high-specific sequence, so DNA is identified as the suitable target for detecting allergens in the high-temperature processed foods. In this study, the PCR assay was established to detect the presence of tropomyosin gene in shrimp and shrimp-derived ingredient foods. In this study, Tropo-F and Tropo-R were successfully designed to detect the presence of tropomyosin gene in shrimp and shrimp-derived ingredient foods. Since tropomyosin is the major allergenic allergen across all shrimps, it was identified as the target molecule in the detection of allergen in shrimp and shrimp-derived ingredient foods [2, 13]. In this study, Penaeus vannamei (white leg shrimp) was used as the positive control for tropomyosin test by PCR assay with designed primers, because it is commonly eaten in Asian countries and is a raw material for shrimp-processed foods [15]. As the results, an 81-bp band, yielded by Tropo-F and Tropo-R, was observed in the sample of Penaeus vannamei (Fig. 2). The accuracy of the assay was confirmed by DNA sequencing, as the result, the signal of sequencing was unique and good for reading, which was homologous to Penaeus vannamei (Accession number: XM_027373346) with homology rate of 100%, and E-value of 0.1 by BLAST (Fig. 3, Table 2). In the negative sample with pork, no band was observed. These results were according to the BLAST results of Tropo-F and Tropo-R primers that only tropomyosin of shellfish, including shrimp, was detected. Additionally, to confirm the applicability of the developed PCR assay, it was initially applied to commercial food samples. As the result, 81-bp bands were detected in three samples of shrimp-derived ingredient processed food. By sequencing, all of these sequences were homologous to the tropomyosin of Penaeus vannamei and Penaeus monodon (Fig. 3, Table 2). These results showed that the designed primer and PCR assay were successful in the development of a DNA-based method to detect the tropomyosin in processed samples due to the more stable property of genomic material. However, the limitation of the current study is less sample enrolled in the study, as well as the sensitivity of the current assay has not been determined. Therefore, further study is necessary to determine the sensitivity of the current PCR assay used in detecting the tropomyosin gene in foods.
Conclusion
Tropo-F and Tropo-R primers were established for detecting the tropomyosin gene. The results confirmed that the PCR assay described and used in the study allows us to detect the food allergen – tropomyosin in shrimp as well as shrimp-derived ingredients in processed food. Further study is necessary to increase the amount of samples as well as to determine the sensitivity of this assay. With further refinement, the set of Tropo-F and Tropo-R could be more useful for the identification and quantitative analysis of the presence of tropomyosin by the development of quantitative real-time PCR.
Acknowledgments
This research was supported by a Grant NO. 04-SVNCKH from Ho Chi Minh City Open University, Ho Chi Minh City, Vietnam.
Declaration of Interest
The authors report no conflicts of interest. The authors are responsible for the content and writing of this article.
References
