Application of SNP Marker For the Identification of Gene Variation Relating to Flowering Time in Soybean
Truong Trong Ngon1, Tran Thi Thanh Thuy2*, Suk-Ha Lee3
|
|
|
ABSTRACT
Flowering at the right time requires the perception and processing of a diverse range of environmental and internal signals. Ninety-five soybean cultivars were used for this study. Eight ESTs relating flowering genes were selected from the soybean cDNA library in NCBI and TIGR. Primer 3 was used to design EST fragments of 500-800bp. Seven ESTs detected 39 different SNP positions. All genes contained exon region except gene Cry 02. Besides, Cop 01 ESTs contained both exon and intron regions. The same ESTs gave different LD among cultivar groups. The LD decay showed that it varied from one cultivar group to another, and from one EST to another. The flowering time of soybean varies a lot from one place to another. Therefore, gene variation at the nucleotide level will be useful for breeders to make a strategy for soybean improvement in the future.
Keywords: Gene diversity; Linkage group; Single Nucleotide Polymorphisms (SNPs).
Introduction
The switch development from vegetative to reproductive stages is the major transition in flowering plants [1-3]. The transition time from vegetative to reproductive phases of the plant is always an interaction between genotype and environment. Flowering depends on the season and growing place and it plays an important role in adaptation to the environment. Thus, the flowering mechanism in higher plants is not only induced by environmental factors like light, and temperature but also controlled by polygenes [4]. Photoperiodism is a factor helping plant response to the length of day and night. Most soybean cultivars belong to the short-day plant group [5]. They flower when photoperiodism is shorter than a critical photoperiod [6]. Cober et al. (1996) [7] reported that there are seven genes affected flowering time and maturity. Under a long day, the dominant alleles suppress and delay flowering time as well as maturity; however, each gene can affect differently [7]. In their study, Tasma & Shoemaker (2003) [8] determined ten gene candidates influencing the flowering time of soybean; these genes were located on ten linkage groups.
Single nucleotide polymorphisms (SNPs) are a useful tool for discovering and exploiting genes more effectively in plant breeding. The nature and frequency of SNP in plants are beginning to receive significant attention. Moreover, SNP markers associated with traits have been discovered in onion, soybean, and rice [9]. Until now, SNP application is useful in plant breeding as it can be automated by a machine called Next Generation Sequence (NGS) [10-12]. The aim of this study was to discover gene variation at the nucleotide level among different soybean cultivar groups.
Materials and Methods
Soybean plant materials
Ninety-five soybean cultivars from Vietnam, China, Japan, and Korea were used to discover SNP in soybean. These cultivars were originated from different ecological regions. They had diverse flowering time. Besides, maturity for each cultivar group was also different. These materials were good for researching gene variation.
DNA extraction
DNA extraction can be carried out by fresh leaves from young seedlings, from 15 to 20 days old, as described by Shure et al. (1983) [13]. DNA concentration was measured by using a Hoechst dye-base protocol for fluorescence spectrophotometer (Model F-4500, Hitachi Lt., Ibaragi, Japan). The DNA solutions were diluted to working concentration with Tris-EDTA buffer (pH 8.0) and stored at –20oC until use.
Design of PCR Primers and Extension Primers
Seven sequences were selected from the soybean cDNA library derived from clones of the NCBI gene bank. One gene of Tentative Consensus sequences (TCs) was chosen from the soybean cDNA library available from The Institute for Genomic Research (TIGR) databases (http://www.tigr.org) (Table 1).
Table 1. Soybean ESTs selected from GeneBank and primer sequence information
|
Accession no. |
Description |
Forward |
Reverse |
|
AI900864 |
COL02 (Constans like 2) |
ATGTCGTATTCGAGTGGGATTG |
GTTCGTACCGATTCGATTCTTC |
|
AI900211 |
GT02 |
GAGACAATGGCTTTGCTCAATA |
GTTGTTGTTGTTGTCGGTTGTT |
|
AW309100 |
CRY02(Cryptochrome2) |
TTGTTTCAACTTTCCCTTCACC |
GGTAGCCATTGCCTCACATATT |
|
AW099970 |
GAI |
ATCCAATAGGTTCGGGCATC |
AAGCATCCTTGATTCTCTTCCA |
|
AI495592 |
CCA1 |
GATCAAATGCTCAAACGGTACT |
AGATGTTCGGTAGAGGCCAAT |
|
AW186405 |
COP1 |
TCCTTGGTGCACATTAGCTG |
AATTGGCACAGGCAGTGATT |
|
AW200732 |
PHYB (Phytochrome B) |
CAGACAAAGCACATGTCACTCA |
GCAGCAAGAAATCAGACAACAC |
Primers were designed using Primer 3 software [14]. The PCR primers were designed to have melting temperatures from 58oC to 68oC, lengths between 20 and 25 bases, and GC content between 40% and 60%.
Initial examination of PCR Primers
All PCR primers were used to amplify the genomic DNA of 95 soybean cultivars. Amplification reaction used standard PCR reagents including 200ng of genomic DNA, 0.8mM of each forward and reverse primer, 200mM of each nucleotide, 10x reaction buffer (750mM Tris-HCl pH 8.5, 200mM (NH4)2SO4, 25mM MgCl2). The total volume of Taq polymerase (Vivagen, Korea) reaction was 50ml. PCR cycling conditions were as follows: 4-minute initial denaturation at 94oC, 30-second denaturation at 94oC, 30-second annealing at 58oC-68oC (depending upon optimal annealing indicated by Primer3), and 1-minute extension at 72oC for 30 cycles on a PTC-225 Peltier Thermal Cycler (MJ Research, Inc. Watertown, MA, USA). PCR products were resolved on 1.0% Ethidium-stained agarose gels. Only PCR products amplified as a single fragment were used for further sequencing reactions. Those produced no or mismatch products were further tested using lower or higher annealing temperature until producing a single amplicon of the predicted length.
Direct Sequencing of PCR products
PCR products producing a single discrete band were purified by AccuPrepâ PCR purification kit (Bioneer, Korea). The purified PCR product was directly sequenced using one of the PCR primers with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystem, Foster City, CA, USA). The reaction reagents including 50ng of template DNA, 0.64mM of primer, 1x reaction buffer (400mM Tris-HCl pH 9.0. 10mM MgCl2), 0.3ml of BigDye Terminator in a total volume of 5ml. PCR cycling conditions were as follows: 4 min initial denaturation at 94oC, 10 seconds denaturation at 90oC, 5 seconds annealing at 50oC, and 1 min extension at 72oC for 40 cycles on an MJ Tetrad thermocycler (MA, USA). The labeling reaction mixture was ethanol-precipitated and resuspended in 10ml of distilled water. ABI 3700 sequencer (Applied Biosystems, Foster City, CA, USA) was used to analyze sequences. Those primers that produced high-quality sequences were used to amplify and sequence the genomic DNA of 95 soybean cultivars as described above.
SNPs Discovery
The sequence data from 95 soybean cultivars were analyzed with SeqScapeâ Software (Applied Biosystem, Foster City, CA, USA). SeqScape is a sequence comparison tool for variant identification, SNP discovery, and validation. It considers alignment depth, the base appears in each of the sequences with base high quality. Putative SNPs were only accepted as true sequence variants if the quality value exceeded 20. It means a 1% chance basecall is incorrect.
Nucleotide Diversity (q )
Nucleotide diversity (q ) was calculated by the method described by Halushka et al. (1999) [15].
q = K/aL a = 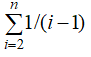
where K is the number of SNPs identified in an alignment length, n is alleles and L is the total length of the sequence (bp).
Results and Discussion
Characterization of SNP marker
The amplified consensus sequence length varied from 270bp to 794bp (Table 2). All SNP markers contained exon, except Col 02. Besides, Cop 01 consists of both exon and intron.
Table 2. Characterization of eight ESTs
|
Genes |
Consensus sequence length |
Sequence region |
No. of SNP |
|
|
|
(bp) |
Exon |
Intron |
|
|
Col 02 |
301 |
1-301 |
|
2 |
|
GT 02 |
359 |
1-359 |
|
3 |
|
Cry 02 |
545 |
|
1-545 |
7 |
|
GAI |
247 |
1-247 |
|
2 |
|
CCA 1 |
270 |
1-270 |
|
1 |
|
Cop 01 |
701 |
559-652 |
1-558; 653-701 |
5 |
|
Phy B |
331 |
1-331 |
|
1 |
Our SNP discovery research in soybean focused on genes designed from EST using 95 cultivars. Exon had the shortest length of 63 bp for Cop 01, and the longest length of 359 bp for GT 02. The more SNP positions, the more diverse in observed genes. The diversity of gene controlling flowering time affected phenotypic variation in the soybean cultivar group because gene function codes RNA messenger and then controls the synthesis of protein.
The diversity of gene controlling flowering time affected phenotypic variation in the soybean cultivar group because gene function codes RNA messenger and then controls the synthesis of protein. Protein and function of seven ESTs related to flowering time were shown in Table 3. The function of eight ESTs responses to light, temperature, and circadian clock regulation, respectively to promote flowering. Of seven EST-encoded proteins, only the protein of two ESTs (GT 2 and Cop 1) was not yet classified (Table 3).
Table 3. Function of proteins encoded by genes related to the flowering time
|
Genes |
Protein encoded |
Function |
References |
|
Col 2 |
Constant like protein |
Promote flowering in response to inductive photoperiod |
Puterill et al., 1995. |
|
GT 2 |
Not yet classified |
Light regulation |
Smalle et al., 1998. |
|
Cry 2 |
Flavoprotein |
Blue-light Photoreceptor |
Lin et al., 1996. |
|
GAI |
Member of a novel family with putative transcription factor |
Promote flowering in response to non-inductive photoperiod |
Peng et al., 1997. |
|
CCA 1 |
MYB-related transcriptor |
Circadian clock Regulation |
Wang & Tobin, 1998. |
|
Cop 1 |
Not yet classified |
Light regulation |
Deng et al., 1992. |
|
Phy B |
Apoprotein |
R-FR light photoreceptor |
Sharrock & Quail, 1989. |
Variation in seven ESTs
Gene variations contained three types: indels, transversions, and transitions, but transversion types were common (Table 4). For each position, the Chinese cultivar group gave many gene variations as compared with the other cultivar groups. Our results contrast with the ratio of transitions to transversions among 95 genotypes. This was similar to the results of Zhu et al. (2003) [16]. A summary of multiple- and single-nucleotide substations, as well as multiple- and single-base indels, is presented in Table 4.
Table 4. Gene variation types of seven ESTs
|
ESTs |
SNP |
Mutant types |
Number of cultivars |
|||||
|
|
position |
|
|
China |
Japan |
Korea |
Vietnam |
Total |
|
Col 02 |
104 |
Transversion |
T |
22 |
15 |
17 |
24 |
78 |
|
|
|
|
A |
2 |
8 |
7 |
0 |
17 |
|
|
267 |
Transition |
A |
22 |
14 |
17 |
24 |
77 |
|
|
|
|
G |
2 |
9 |
7 |
0 |
18 |
|
GT 02 |
240 |
Transition |
G |
24 |
22 |
23 |
24 |
93 |
|
|
|
|
A |
0 |
0 |
1 |
0 |
1 |
|
|
284 |
Transversion |
G |
0 |
0 |
1 |
0 |
1 |
|
|
|
|
T |
24 |
22 |
23 |
24 |
93 |
|
|
290 |
Transversion |
C |
24 |
22 |
23 |
24 |
93 |
|
|
|
|
A |
0 |
0 |
1 |
0 |
1 |
|
Cry 02 |
137 |
Insertion |
A |
23 |
23 |
24 |
24 |
94 |
|
|
|
Deletion |
- |
1 |
0 |
0 |
0 |
1 |
|
|
146;1- |
Insertion |
CTTCT |
2 |
0 |
0 |
6 |
2 |
|
|
146;5 |
Deletion |
----- |
22 |
23 |
24 |
18 |
87 |
|
|
193 |
Transversion |
A |
24 |
21 |
23 |
24 |
92 |
|
|
|
|
C |
0 |
2 |
1 |
0 |
3 |
|
|
211 |
Transversion |
A |
23 |
23 |
24 |
24 |
94 |
|
|
|
|
C |
1 |
0 |
0 |
0 |
1 |
|
|
268 |
Transversion |
A |
23 |
23 |
24 |
24 |
94 |
|
|
|
|
T |
1 |
0 |
0 |
0 |
1 |
|
|
298 |
Transversion |
G |
23 |
23 |
24 |
24 |
94 |
|
|
|
|
C |
1 |
0 |
0 |
0 |
1 |
|
|
320 |
Transition |
G |
23 |
23 |
24 |
24 |
94 |
|
|
|
|
A |
1 |
0 |
0 |
0 |
1 |
|
GAI |
229 |
Transition |
G |
24 |
16 |
18 |
21 |
79 |
|
|
|
|
A |
0 |
6 |
6 |
1 |
13 |
|
CCA 1 |
69:1 |
Insertion |
A |
1 |
1 |
1 |
1 |
4 |
|
|
|
Deletion |
- |
23 |
22 |
23 |
21 |
89 |
|
|
150 |
Transition |
G |
20 |
23 |
23 |
22 |
88 |
|
|
|
|
A |
4 |
0 |
1 |
0 |
5 |
|
Cop 01 |
266 |
Tranversion |
T |
22 |
23 |
21 |
20 |
86 |
|
|
|
|
A |
2 |
0 |
2 |
4 |
8 |
|
|
468 |
Transition |
T |
22 |
23 |
21 |
20 |
86 |
|
|
|
|
C |
2 |
0 |
2 |
4 |
8 |
|
|
516 |
Transversion |
T |
0 |
1 |
0 |
5 |
6 |
|
|
|
|
A |
24 |
22 |
23 |
19 |
88 |
|
|
530 |
Transversion |
C |
22 |
23 |
21 |
21 |
87 |
|
|
|
|
A |
2 |
0 |
2 |
3 |
7 |
|
|
588 |
Transition |
G |
22 |
23 |
21 |
20 |
86 |
|
|
|
|
T |
2 |
0 |
2 |
4 |
8 |
|
Phy B |
254 |
Insertion |
AG |
24 |
23 |
24 |
22 |
93 |
|
|
Deletion |
-- |
0 |
0 |
0 |
2 |
2 |
|
Seven observed ESTs detected about 39 SNP. Among them, Cry 02 gave high SNP positions (Table 4). Chinese, Japanese, and Korean cultivar groups had more variation than the Vietnamese cultivar group through seven ESTs observed. This demonstrated that the Vietnamese cultivar group had narrow genetic variation. This indicated that selection affected the Vietnamese cultivar group, or the mutation rate was low. It is difficult to get a genetic advance with a narrow genetic variation source in plant breeding.
DNA Polymorphism at seven ESTs
Nucleotide polymorphism can be measured by many methods, for example, haplotype (gene) diversity, nucleotide diversity, Pi (p) [17], Theta (q) (per site), etc.
In our study, nucleotide diversity was estimated as theta (q), the number of segregating sites [18], and its standard deviation (Sq). These parameters were estimated by DNA Sequence polymorphism software version 4.0 [19]. Theta value explained the nucleotide diversity of sequences for each gene. The higher the values, the more the diversity. Chinese cultivar group had the highest theta values in Col 02, GAI, and Cop 01. Other cultivar groups gave high theta values one ESTs, for example, the Japanese group for Cry 02, Korean group for GT 02, and Vietnamese group for CCA 1 (Table 5). Chinese cultivar group had the highest theta values in three of the eight markers (Table 5).
Table 5. Nucleotide diversity (q) values of eight ESTs using the program DnaSP 4.0 (Rozas et al. 2005)
|
ESTs |
China |
Japan |
Korea |
Vietnam |
|
Col 02 |
0.677 |
0.659 |
0.651 |
0.556 |
|
GT 02 |
0.323 |
0.533 |
0.630 |
0.610 |
|
Cry 02 |
0.589 |
0.622 |
0.554 |
0.614 |
|
GAI |
0.559 |
0.463 |
0.370 |
0.462 |
|
CCA 1 |
0.532 |
0.625 |
0.661 |
0.720 |
|
Cop 01 |
0.561 |
0.449 |
0.516 |
0.480 |
|
Phy B |
0.309 |
0.511 |
0.428 |
0.436 |
|
Mean ± SD |
0.518 ± 0.13 |
0.556 ± 0.08 |
0.549 ± 0.11 |
0.542 ± 0.10 |
|
CV% |
25.46 |
14.12 |
19.27 |
18.48 |
In summary, one of the most important environmental factors affecting flowering time is the daily duration of light, the photoperiod, which was first discovered by Garner & Allard in the 1920s [18]. Recent molecular genetic studies in a facultative long-day plant, Arabidopsis, have made notable progress in identifying genetic pathways and molecular components associated with the control of flowering time and the function of the circadian clock, which have been discussed in two recent Updates [20, 21]. From Arabidopsis alone, at least 80 loci were reported to affect the timing of flowering [22]. For soybean, at least seven genes have been reported to affect flowering time and maturity [7]. In our study, eight ESTs detected 39 SNPs. Chinese cultivar gave more diverse than others. Vietnamese group had a narrow variation. SNPs are the only new generation molecular markers for individual genotyping needed for molecular marker-assisted selection (MAS). There is some evidence that the stability of SNPs and, therefore, the fidelity of their inheritance is higher than that of the other marker systems like SSRs and AFLPs [9].
Conclusions
Seven observed ESTs detected about 39 SNP. Among them, Cry 02 gave high SNP positions. Chinese, Japanese, and Korean cultivar groups had more variation compare to the Vietnamese cultivar group through seven observed ESTs. This demonstrated that the Vietnamese cultivar group had narrow genetic variation. This indicated that selection affected the Vietnamese cultivar group, or the mutation rate was low. Chinese cultivar groups were more diverse than other groups. All observed genes contained exon region; except Cry 02. Furthermore, Cop 01 genes contained both exon and intron regions. With same primer different LDs among cultivar groups were observed. The longer distance, the lower r2. In conclusions, there was wide variation in flowering time of soybean. Gene diversity at nucleotide level will be useful for breeders to make strategy for soybean improvements in the future.
References
