The Importance of The Environment in The Study of Metagenomics and Microbiome
Bruno Riccardi 1*, Sergio Resta 2
|
|
|
ABSTRACT
In recent years, the study of the microbiota has focused the attention of research through the correct interpretation of the set of microorganisms present in various organic surfaces of different animal species and humans, and on the “Metagenome”, which is the study of the DNA sequences of the microbiota. The researchers' attention was focused in particular on demonstrating the close relationship between the variation of the microbiota and the effects it produces on the state of health or on the causes of various pathologies. This scientific document is a review of the most recent literature on the topic "microbiome and state of health" with the aim of restoring the cardinal role of the genetic, environmental, and dietary variables that alter both our intestinal homeostasis and our health.
Keywords: microbiota, microbiome, metagenetics, intestinal homeostasis, microbial dysbiosis.
Introduction
The powerful influence that the environment has on living beings has been known for centuries and the work of Charles Darwin "On the origin of species" has masterfully documented the fundamental role that the environment plays in the selection and natural evolution of life [1].
In an ecosystem, there are always two physical quantities, which in addition to exerting an effect of mutual transformation between them, at the same time modify the environment in which they operate: Energy and Matter [2], which are schematically represented in the following diagram Fig.1
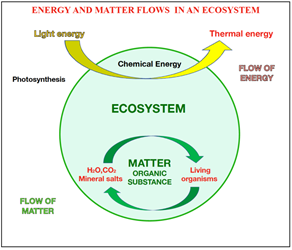
Fig. 1: Energy and matter flows
The flow and mutual effect between matter and energy, which represent the two sides of the same coin (there is no energy without matter), have had a significant impact on all ecosystems.
In millions of years of evolution, the action between matter (in particular organic matter, biomass) and energy has shaped: archaea, bacteria, plants, animals, and man and all the countless species that live on our planet and share the same ecosystem [3].
The closer the link between the different species that coexist in the same environment, the greater the influence they exert on each other, and, reciprocally, the effect that these biological entities determine on the surrounding environment.
This type of influence is very evident if we analyze the different ecosystems.
A new specialization in Ecology has been developed for some years now, centering its focus on the study of all microorganisms (Microbiota) present in the internal and external environment of the organism of various living species, including the human being. Equal and contemporary doctrinal development has had the Microbiome, which is the study of the genetic heritage attributable to the microbiota. Two compartments of the same material, one functional to the other. [4-10]
The microbiota in mammals and humans
Mammals, like other vertebrates, live in co-evolutionary association with the plethora of microorganisms present in a variety of different microenvironments.
The microbiome study has developed exponentially in recent years. Thousands of scientific works have been produced and its exploration is constantly evolving. International associations have sprung up, specialized magazines for the study dedicated to this prolific new topic. [6]
Thousands of microorganisms, bacteria, yeasts, viruses, etc. have been studied with different investigation methods: biochemistry, metabolic, evolutionary, taxonomic in order to examine the effects they produce on host organisms [11].
Furthermore, modern genetic sequencing technology has made available for research effective tools for the typing of microbial species present in different ecosystems, through the study of DNA and the interpretation of the ribosomal operon, in particular of the gene that codes for rRNA 16S. (The latter is the component of the small 30S subunit of a prokaryotic ribosome that binds to the Shine-Dalgarno sequence. The genes that encode it are referred to as the 16S rRNA gene and are used in the reconstruction of phylogenies, due to slow evolution rates of this region of the gene). [12, 13]
The choice of the use of the 16S rRNA gene as a phylogenetic marker to examine the diversity of the microbiome and to identify and classify microorganisms derives from the difficulty in cultivating most of the microorganisms present in natural environments.
The bacterial microbiota thrives on practically any surface of the human body, but the most colonized organ is the gastrointestinal tract in which the quantity and diversity of the microbiota progressively increase from the stomach to the small intestine and colon; only in the large intestine is present over 70% of all microorganisms present in the human organism: it is an environment dominated by anaerobic bacteria, in particular, the close one, as well as by viruses, protozoa, archaea, and fungi.
From these studies, a map derives that represents the different distribution of various microbial species that make up the microbiota in our gastrointestinal system. Fig.2
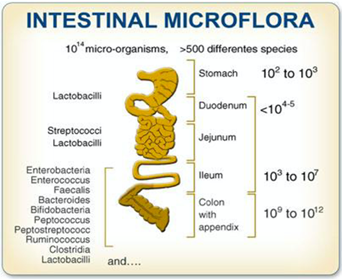
Fig. 2: Composition of the gut microbiota from a space-time point of view
Further development of research on the microbiota led to the birth of Metagenomics, which is the study of DNA sequences belonging to the biological entities making up the biota itself.
A metagenomic analysis is based on the study of a series of DNA sequences of different microorganisms and is very useful when some of them are difficult if not impossible to grow.
To determine the genetic identity of an organism (genome) or a set of organisms (metagenome) it is necessary to determine the sequence of all DNA molecules present in the sample. [13, 14]
The human organism is an extremely complex environment. Our body is made up of about 1013 different cells, but it contains ten times more (about 1014) commensal microorganisms, the so-called "human microbiome", which has a profound influence on physiology, nutrition and is crucial for our health.
It provides nutrients and vitamins, supports the response to infections, and actively eliminates various toxic substances.
Metagenomics today makes it possible to characterize the composition and dynamics of the population of the human microbial community and the complex cooperative (or antagonistic) interactions with human cells and tissues.
Microbiome variability, cause, or effect of pathologies?
The altered functionality of the human microbiota (dysbiosis) compared to the normal intestinal flora (Fig. 3) would seem to play, according to many studies object of this review, a key role in various pathologies such as obesity, cardiovascular diseases, chronic inflammations, intestinal diseases, metabolic syndrome, diabetes, various localized infections, neuronal diseases, cancer, etc. (the list is constantly expanding).
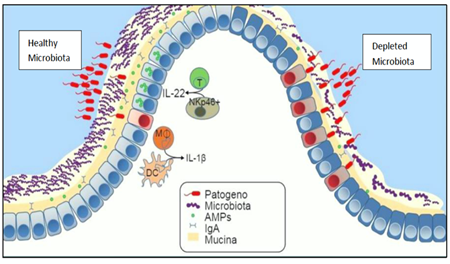
Fig. 3: Comparison between normal microbiota and intestinal dysbiosis
A solid causal relationship between some bacterial or viral strains or parasites that invade the intestine and the related gastrointestinal diseases generated by them is an acquired fact. Bacterial species: Salmonella, Shigella, Campylobacter, Clostridium, Escherichia coli, Vibrio cholera, viruses such as: rotaviruses, noroviruses, adenoviruses, coronaviruses, and parasites: giardia lamblia, cryptosporidium, entamoeba histolytica are among the most frequently associated pathogens.
But attributing a relationship between the variability of the microbiome (meaning its extreme biological dynamism) and the different organic pathologies is at least questionable. This vision in some ways "displacing" the effects of the microbiome on human health, obscures the beneficial effects that it undoubtedly has on our body. [1, 15, 16]
Intestinal flora has been shown to have many beneficial functions, such as the synthesis of vitamins K, B12, B1, B6, B9, and B2, digestion of dietary fiber, and regulation of inflammatory responses.
Equally evident is the great variability of its composition in bacteria, yeasts, viruses, fungi inextricably linked to the diet and other external factors, capable of interacting and modifying the environment that hosts it. [17]
Since the microbiome depends on the variability of different environmental factors (defined as "dependent factors"), it cannot be considered as the only trigger for all the diseases attributed to it by the reported studies. Except, of course, for most gastrointestinal ones, generated by specific bacterial, viral, and parasitic strains.
As for the numerous published studies, it is not always possible to grasp a rigorous objective in their method, since they are often spoiled by unilateral interpretations and conclusions. At this point, it is appropriate to remember the importance of the scientific method that each Author should follow.
The research path is based on experimental platforms, it is the typical way in which science proceeds to obtain a knowledge of objective, reliable, verifiable, reproducible, and shareable reality.
Good clinical practice (GCP) or better, good clinical practice guidelines, were adopted by the European Union in 1996 and transposed in Italy with Ministerial Decree on July 15, 1997, as follows:
"Good clinical practice is an international standard of ethics and scientific quality for the design, conduct, recording, and communication of clinical studies involving human subjects. Adherence to these GCP standards publicly guarantees not only the protection of rights, safety, and the well-being of the parties under study, in accordance with the principles established by the Helsinki Declaration, but also the reliability of the study data. "International Conference on Harmonization (ICH) - European Medicines Agency (EMA) 1996”
Materials and Methods
This study is a meta-analysis of the most significant scientific literature recently produced on the topic "microbiome, its balance and state of health" and aims to shed light on the many variables that alter this balance, magnifying the fundamental role that the microbiome plays for our health. [17-20]
Overall, a large part of the international literature is excessively exemplified, often absolutely not credible, leading to misleading pathogenetic conclusions on the role of the microbiota, considered to be the only protagonist and creator, "Deus ex machina", of the various nosographic processes described. As a demonstration of the interpretations supported by the vast number of publications on this topic, only a few articles are indicated here (it is impossible to examine them all for their relevant number) which can be summarized on an argumentative basis:
A) Effects of the microbiota on cardiovascular diseases:
According to the articles reported, these would be exclusively connected to the presence of circulating trimethylamine N-oxide (TMAO), a typical product of intestinal dysbiosis. The studies describe from an epidemiological point of view the multiple primary risk factors known for these diseases (hypertension, total hypercholesterolemia, and LDL, obesity, hyperhomocysteinemia, etc.) but according to some, they assume a secondary role compared to the circulating TMAO produced by the abnormal microbiome, considered - reductive - as the real cause of cardiovascular diseases. [21-24]
B) Effects of the microbiota on intestinal and metabolic diseases:
The dedicated articles highlight the interaction between diet, lifestyle, and microbiota, but only the latter is described as the cause of various intestinal and metabolic diseases. We, therefore, recommend the use of probiotics, prebiotics, and symbiotics in the prevention of important enteropathies such as irritable bowel syndrome, gastritis and reflux disease, obesity, diarrhea, inflammatory bowel disease (IBD), ulcerative colitis (UC), disease Crohn's, cancer, etc. when, on the other hand, we know how many other etiopathogenetic "movens" are at stake in numerous pathologies of the small and large intestine. [15, 25-28].
C) Variability of the microbiota in relation to the environment and the host:
An examination of the literature shows that the potential pathogenic role of the microbiota is irreplaceable for the host organism and that the other variables that come into play and alter the state of health are not taken into consideration, if only marginally.[16]
D) Diet is one of the main independent variables that can influence the gastrointestinal ecosystem.
In the description of intestinal physiology and pathophysiology, a prominent position must be attributed to it, because very often it is unbalanced in the qualitative and quantitative supply of nutrients, but above all, because it can be a pathogenic microbial loading vehicle. In reality, food and drink are never sterile, but on the contrary, they always contain pathogens that can generate toxins, very frequent for example in the incongruous diets of underdeveloped countries. Not to mention pesticides and chemical additives, GMOs, disturbing and unrecognized presences. Another important variable is the presence of genetic defects in the host that are metabolic pathologies, which can have serious effects on intestinal health and the general well-being of patients. However, no study refers to the direct contribution of the plasma metabolites originated by the microbiota, to the intrinsic variables linked to this activity dependent on the food intake. (Fig.4)
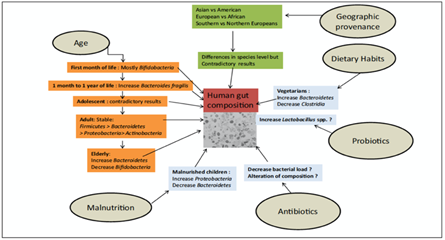
Fig. 4: The influence of external factors determining the composition of the human gut microbiota.
The recent Sars-Cov2 virus has brought to our attention some of the feasible causes in terms of viral contamination due to failure to comply with common hygiene measures (hand cleansing, interpersonal contacts maintained without particular precautions, contaminating sputum, orofecal transmission, etc.). What consequences do these factors have on the composition of the gut microbiome? All these vehicles of infections, present in the daily life of each individual, can influence and alter our microbiome at any level. The last but not least effect is the possible contamination during the collection of the examined samples, which can influence the results of the metagenomic investigation and influence their interpretation.
So the variability of the intestinal ecosystem can depend on several factors:
Physiological factor: inadequate diet and age of the subjects observed, with important qualitative and quantitative changes on the microbiome: for example, in young people, lactobacteria reach high concentrations, while in the elderly there is a significant reduction in bifidobacteria and an increase in coliforms and fungi. (Fig.5)
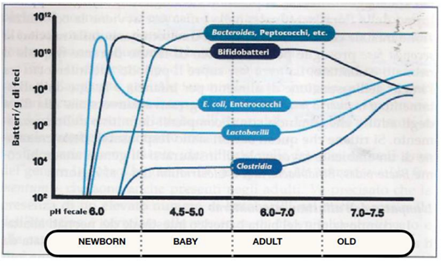
Fig. 5: Variation in concentrations of bacterial colonies of the microbiota in relation to age
Pathological factor: acute and chronic intestinal diseases, systemic diseases, hormonal alterations, stress, immunological diseases, liver disease, organic degeneration at various locations, cancer;
Iatrogenic Etiology: surgical procedures, antibiotics, analgesics, and allopathic antiphlogistic pharmacopeia,
Chemical additives: present in animal and plant foods (pollutants capable of significant changes in the microbiome as well as a real procarcinogenic action).
There is an obvious change in the composition of the microbiome during the evolution of various human ages. From pediatric to mature age the microbiome is in constant qualitative-quantitative modification, should we, therefore, conclude that this variation is the cause of aging? Fig.5
Finally, what value should we attribute to the infinite scientific publications, clinical studies, cohort studies that have investigated and continue to examine the various pathogenetic causes and risk factors for the same diseases that today we attribute univocally and arbitrarily to the variation of the microbiome?
Do we still have to consider them valid scientific references or should we relegate them to the acquisitions surpassed by the most recent metagenomic research?
Why so much attention to the intestinal microbiome only?
The endless production of research on the intestinal microbiome contrasts with the reduced production of studies on other ecosystems such as the vaginal, respiratory, urogenital, skin, etc. This is due to the complexity and importance that the intestine occupies in the state of well-being and disease of our organism.
There remains the problem of the difficult understanding of the causes of many diseases that affect the human species, in particular the chronic-degenerative ones.
A fruitful path may be to adopt a "holistic" and strictly scientific approach to the study of the most common diseases, which takes into account, also - but not only - the microbiome and all the other variables that come into play. [29]
The study of the microbiome that does not take into account all the variables indicated, all of which have a significant impact on its composition, is a sophisticated intellectual exercise even if ennobled by the high technological content of the analytical tools used. It is necessary to attempt an ideological orientation free from partial contamination in order to give the right etiopathogenetic recognition to all those factors capable of breaking the subtle balance of the microbiome in conjunction with congenital structural alterations, dysmetabolisms, genetic modifications, pathological events capable, by themselves, of arousing disease and objective symptoms.
Conclusions
In recent years, there has been a growing interest in the study of the microbiota, the set of microorganisms present in various animal species and in man. The attention of the researchers has been focused in particular on the investigations related to the characteristics of the human intestinal flora and on the effects that the modifications of the microbial population produce on the state of health or in the determinism of various pathologies.
Although these studies have contributed greatly to the knowledge of the intestinal ecosystem and gastroenteric pathophysiology, they have also attached too much importance to the role of the microbiota in the pathogenesis of an increasing number of diseases.
Disclosure
The authors report no conflicts of interest in this work.
References
